User Manual for
Ana
Michael Pusch, Istituto di Biofisica, Genova, email:
pusch@ge.ibf.cnr.it
Last change: 23.08.2007
Index
1. Introduction
1.1. Installation
2. Subwindows
2.1. Tree window
2.2. Traces window
2.2.1. Filtering traces
2.3. Results window
3. The relevant segment
4. Cursors in the "trace" window
5. Short-cut keys
6. Scaling the display of the traces
7. Parameter settings
7.1. Editing parameters
7.2. Loading/Saving
7.3. Context sensitive parameters
7.4. Undoing a parameter settings change
8. Editing comands - The internal buffer
8.1. Undoing an erroneous operation :
Exchange
8.2. Copying/pasting/cutting/deleting/merging traces
and series
8.3. Permanent filtering
8.4. Undersampling
8.5. Calculating the average
8.6. Copying selected records
8.7. Concatenate entries
8.8. Deleting a stretch
8.9. Extracting a stretch
8.10. Overlay envelope
8.11. Swapping data and fit
8.12. Reverse time
8.13. Paste as fit
9. Trace fitting
9.1. Exponential fits
9.1.1. Simple exponential fit
9.1.2. Fit several traces
simultaneously
with the same time-constants for all
9.1.3. Other exponentials
9.1.4. Envelope
9.2. Polynomial
9.3. User defined
10. Series Fit
10.1. Boltzmann
10.2. Polynomial
10.3. User defined function
11. Leak, capacity, and line frequency
(50/60
Hz) subtraction
11.1. Baseline subtraction
11.2. Leak subtraction
11.2.1. subtract a scaled series
11.2.2. Subtract using a baseline
segment where channels are supposed to be closed
11.3. Capacity correction
11.4. line frequency (50/60 Hz) subtraction
11.4.1 Automatic line frequency (50/60 Hz)
subtraction
11.4.2 Manual line frequency (50/60 Hz)
subtraction
12. Scaling traces
12.1 Scaling by a factor and add
a constant
12.2 Scaling traces of a series to have a
common
value in a certain segment
13. Noise analysis
13.1. Selection of records and cursor settings
13.2. Variance-mean analysis
14. Single channel analysis
14.1. Amplitude Histogram and Gaussian
Fit
14.2. Idealization
14.3. Manual editing of the idealized record
14.4. Dwell time analysis
14.5. Binomial Peak analysis
15. Spectral analysis
16. Exporting data
16.1. Export raw data
16.2. Export analysis results (mean, tau,
peak, etc)
16.3. Export noise analysis
16.4. Export spectrum
16.5. Export amplitude histogram
16.5. Export dwell time histogram
17. Misc.
17.1. Changing the pulse protocol ("Stimwave")
18. Appendix
18.1. Supported data file types
18.2. The internal data-format of ana: what is
saved on a file?
18.3. Syntactic rules for the user-defined function
1. Introduction
This manual is contained in only one html-file to allow the use of the
Find
feature of your browser in the whole manual and to facilitate its
printing.
Ana is a program for the analysis of
electrophysiological
data. The emphasis is on pulsed data, in particular of
voltage-dependent
currents. But also continuous data can be analyzed. In particular single-channel
analysis is possible.
Ana can read various file-formats
.
Data can be
edited extensively (i.e.
traces or parts of traces can be deleted, copied, pasted, ...).
The main purpose is
fitting of traces,
extracting
of IV´s (and other things like that),
leak
subtraction ,
scaling,
(non-stationary)
noise analysis,
single-channel analysis,
and
spectral analysis.
Ana is not able to save data in any of the
formats
used by the standard acquisition programs but ONLY in its own
format! In this format, however, it is possible to store a lot of
data
analysis.
The program is for free but no responsibility whatsoever is assumed
by the author. For the latest version, comments and suggestions
contact:
Michael Pusch, Istituto di Biofisica, Genova, email: pusch@ge.ibf.cnr.it.
Known limitations and bugs:
- The complete data file is loaded into memory. Its size must
therefore
not exceed the memory of the computer.
- Practically no internal checks are done for sufficient memory. This
may lead to crashes if more memory is needed than available.
- Sometimes the program may crash if it is attempted to read a data
file in a format that is not supported.
1.1. Installation
No special installation or registration is needed. Ana
is contained in the single ana.exe file that can be launched
from
where you want. It might be convenient to have a pool of setting
files (*.set) in a dedicated folder.
2. Subwindows
More than 1 file can be loaded at the same time. The window for each
file
is divided into 3 subwindows. The left subwindow
is
mainly for a tree-like view of all traces of the file allowing their
simple
selection. The right part of each window is divided into 2 parts. In
the
lower
part the currently selected data traces are plotted. In the
upper
half various results of previous analysis or other information is
displayed.
2.1. Tree window
The tree window shows in the upper part an expandable tree structure of
the data. The hirarchy is: Root->Series->Entry.
Each series is composed of several "entries". A file may contain many
series'. The series' are numbered sequentially, starting from 1. Also
the
entries are numbered sequentially, and the entry numbers DO NOT START
WITH
"1" in each series. You can move within the tree using the arrow keys
or
using the mouse.
For displaying a single entry or a whole
series,
the following rules apply:
If you have selected Update on moving in tree in the Settings-dialog
SINGLE ENTRIES are displayed in the trace window
whenever the cursor is on them. Otherwise, single entries, are
displayed
only if you hit return or if you click the RIGHT mouse
button.
The latter rule applies ALWAYS for whole series'.
The controls below the tree are explained in separate sections (i.e.
the
sliders and the relevant
segment).
2.2. Traces window
Here the actual, currently selected , data
traces
are normally displayed. In addition five vertical and two horizontal
cursors
and a zero line are displayed. The time- and y-range that are displayed
are changed as described in 6. Scaling the
display
of the traces.
The display and/or subtraction of leak-records ("P/N") is controlled
as described in the
7. Parameter settings
section.
If you have performed noise analysis
you
can display the mean and the variance in the trace window by selecting
View->LowerWindow->Noise.
If you have performed spectral analysis
you can display the spectrum in the trace window by selecting View->LowerWindow->Spectrum.
2.2.1. Filtering traces
A Gaussian filter can be activated by inserting the desired frequency
value
(in Hz) in the corresponding field in the
parameter
settings dialog. Filtering does NOT change the data, but is only
for
display.
If you want to filter and change the data permanently, see 8.3.
Permanent filtering.
For undoing such filtering, see 8.1.
Undoing
an erroneous operation : Exchange.
2.3. Results window
The upper right window can display various kind of information. The
actual
viewtype is selected in the menu point View (or via the
toolbar).
If View->General is selected the following general
information
is displayed:
If the root in the tree window is selected,
general
information like filename, number of series and entries etc. are shown.
If a series in the tree window is selected,
information
about this series is displayed, including sample time, number of
entries,
gain, and a list in which for each entry is shown:
i) index
ii) the actual "x-value" of this segment. This depends on the relevant
segment and is either a voltage, a duration of the segment, the
time-point
of acquisition of this entry or its 0-based index. The "x-value" is
used
for export of analysis results and for the series
fit
iii) the mean current calculated between the 2 green
cursors in the "trace" window
If View->SeriesFit is selected the results of the series
fit are displayed. A common example of this is an 'I-V'.
If View->StimWave is selected the pulse program that
generated
the data is shown as a table. The time and voltage values of the pulse
program are used by some procedures to locate cursor
positions
and they are used by some leak subtraction procedures.
The pulse program is also very important for the definition of the relevant
segment.
If View->Analysis Results Table
is selected, a table of analysis results of the currently selected
series
is displayed. The first column contains the respective "x-value" that
is
determined by the relevant segment and
is
either a voltage, a duration of the segment, the time-point of
acquisition
of this entry or its 0-based index. The other columns are speciefied in
the settings dialog and are values
like
initial value, mean, time constants etc. Importantly, the text of the
table
can be selected with the mouse and copied into the windows clipboard
with
a right button mouse click. Note that selecting Select All in
the popup menu that pops up with a right button click AUTOMATICALLY
ALSO
COPIES EVERYTHING INTO THE WINDOWS CLIPBOARD.
If View->Stimwave is selected, the pulse protocol of
the currently
selected series is displayed. If you wish you can change it with the Edit->Stimwave
command. See 17.1. Changing the pulse protocol
("Stimwave")
If View->ShowSomePoints is selected, data points around
the
left green cursor (left column) and the right
green
cursor
(right column)are shown of the currently selected entry are shown. The
values are the zero-based index and the current in pA. See 4.
Cursors in the "trace" window how to move the cursors. The arrows
indicate
the position of the respective cursor.
If View->AmplitudeHistogram is selected, the all-points
histogram
is displayed if this has been constructed previously (see 14.1.
Amplitude Histogram and Gaussian Fit)
If View->BetaDistributionFit is selected,...
If View->LorenzFit is selected, a log-log plot of the
power
spectrum together with a fit with the sum of Lorentzians is shown, if
it
has been previously constructed (see 15.
Spectral
analysis).
If View->DwellTimeHistos is selected, log-plots of
cumulative
dwell-time histograms of the various single channel conductance levels
are shown if these have been previously constructed (see 14.3.
Dwell time analysis).
If View->VarMean is selected, the variance-mean plot of
the
non-stationary noise analysis together with a parabolic fit is shown if
it has been previously constructed (see 13.
Noise
analysis).
3. The relevant segment
In an IV-plot you plot current versus voltage. The relevant segment
serves
to "tell" the program which voltage shall be used for the IV-plot. It
is
taken from the voltage-protocol that was used to generate the series
(see
2.3.
Results window to get info on the voltage-protocol).
Note that counting starts with zero, i.e. the first segment has index
0.
The relevant segment is selected either in the control in the lower
part of the Tree window or in the Parameter
settings dialog.
If you do not want to use the voltage of a segment as the "x-value"
but rather its duration as would be the case in "recovery from
inactivation"
protocols, you have to uncheck the Segment Voltage (else: segment
duration)
box in the Parameter settings.
If you want to analyze the series data not as a function of voltage
or segment duration but rather as a function of the absolute time of
the
acquisition of the entries or as a function of the index within the
series,
you have to select "-2" or "-1" respectively as the relevant segment.
4. Cursors in the "trace" window
There are 5 vertical (2 green, 2 red and one blue) and two (red)
horizontal
cursors in the Traces window. One of each group
(i.e.
one vertical and one horizontal) cursor is SELECTED, implying that its
color is gray and that its movement can be controlled by the keyboard
(see
below). You can toggle with the SPACE bar the currently
SELECTED
cursor.
Vertical cursors are moved by the mouse (LEFT BUTTON!) and the selected
(gray) cursor can additionally be moved with the LEFT and RIGHT arrow
keys
in combination with either the SHIFT key (fine movement) or the CONTROL
key (coarse movement) (see 5. Short-cut keys
for
an overview of frequently used key-commands).
Attention: if you try to move a cursor by dragging with the left mouse
button it may happen that you don't grasp it correctly and that you
instead
select a small display range (or vice-versa) (see 6.
Scaling the display of the traces). The best thing to do in that
case
is to press Ctrl+Z in order to undo the display scale setting
(see 7.4. Undoing a parameter settings change).
Horizontal cursors are moved with the right mouse button and the
SELECTED
(gray) horizontal cursor can additionally be moved by
SHIFT+ARROWUP(DOWN)
(fine movement) and PAGE-UP(DOWN) (coarse movement)(see 5.
Short-cut keys for an overview of frequently used key-commands).
The horizontal cursors serve (so far) for only one purpose: to
measure
amplitudes of any kind "by eye". The difference between the two
corresponding
values of the horizontal cursors is displayed as "cursors" if View->General
is currently selected and if a single entry is displayed. (see 2.3.
Results window).
5. Short-cut keys
Many shortcuts are found in the menus. Since Sept. 2005 Ctrl+C or Edit->Copy (Copy) has a new
behavior in certain situations: If in the upper right window (the results-window) text in a gray results-field
is selected, and if the results-window is active (i.e. the selection is
visible), the short-cut "Ctrl+C"
(or Edit->Copy) copies
this text into the general Windows clipboard. It will not copy the
selected tree item into the buffer as is
instead normally done by the Copy command.
Some shortcuts are listed below:
Cursor movement: With space
the
active (gray) cursor is toggled
Move the selected (gray) vertical cursor with
Shift+Left/right
arrow for a fine movement and with Ctrl+left/right arrow
for a coarse movement.
Alt+LeftArrow(RightArrow): Move ALL cursors
corresponding to the time increment of the relevant stimulation segment
Shift+Arrowup(Arrowdown): Move selected horizontal cursor
by about one pixel
PgUp(PgDown): Move selected horizontal cursor
by about 50 pixel
Data Editing: Arrow keys: move in the tree (if tree
sub-window
is active);
Right mouse click or return in the data tree: activate the
selected item for display and analysis;
Misc.: Plus (minus) key on keypad: Double (reduce to half)
display
gain
Ctrl+PgUp(Down): Shift the traces up/down
With del the currently selected item of the tree window is
deleted; see 8.2.
Copying/pasting/cutting/deleting/merging
traces and series
6. Scaling the display of the traces
The traces can be scaled up/down by the +/- key of the
keypad,
or by the corresponding toolbar buttons. The display gain can be
defined
also in the Parameter settings
dialog.
Traces can be shifted up or down by check Ctrl+PgUp/PgDown or
by the arrow buttons in the toolbar or by defining the value of the
y-offset
in the Parameter settings dialog.
The visualized time-range can be selected by the slider controls of
the Tree window. It can be scrolled through also
with
these controls. The time-range can also be defined in the Parameter
settings dialog.
An easy way to select a rectangle for enlargement is to define it
directly
with the mouse (keeping the left mouse button pressed). Since also the
vertical cursors are moved with the left mouse
button
pressed you have to make sure that the starting edge of the rectangle
does
not overlap with a vertical cursor.
It may happen that you erroneously selected a too small region and
you would like to return to the previous display settings: just hit Ctrl+Z
(or the undo button of the toolbar) . A circular buffer keeps the last
20 display (and all other) settings in memory. With Shift+Z
you
can redo the changes in the settings. See also the menu items Edit->Undo
and Edit->Redo.
7. Parameter
settings
Several parameters that concern
display settings
,
cursor
positions , definition of fit functions, many switches and other
parameters
are kept in the Parameter Settings structure that can be saved in a
settings
file (.set) and reloaded in several ways. The last settings file that
has
been stored by ana is automatically loaded at the next execution.
7.1. Editing parameters
The parameters are accessible in various ways:
Display parameters are most commonly changed as described in
display
settings .
Cursor positions are determined as described in
cursor
positions .
The "General Parameters" dialog is recalled by Settings->General,
or by Ctrl+A or by the corresponding toolbar button. Here you
can change:
- the display gain, Offset x (left and right) (for the time range),
Offset Y (see also display settings );
- the relevant segment;
- the "noise margin" (times SD): this value is used in the
selection of records in the
noise analysis.
The higher the value, the more tolerant is the automatic procedure for
the rejection of "bad" traces;
- the "Scale Var Trace": if you display in the trace
window the result of a noise analysis
this parameter is used to scale the display gain of the variance trace;
- "SD Gaussian noise / pA": with the command Edit->rarely
used->Add white noise gaussian noise can be added to the
traces. The
SD of this added noise (in pA) is defined here;
- "number of components": this value determines the number
of terms for various kinds of fits: For
exponential
fits it determines the number of exponential components. For polynomial
fits it determines the number of coefficients;
- "Filter (Hz)": this value defines the value used for the
digital Gaussian filter. A value of 0 dictates
that
no filtering is to be done;
- "ABF gain factor": When reading Axon Binary Format files,
there was sometimes a problem with the gain. The value defined in this
field is used to multiply the gain found in the ABF file, and serves to
get correct current values. This parameter is now obsolete. See also 18.1.
Supported
data file types;
- "Line Frequency": If you use the manual
line frequency (50/60 Hz) subtraction at this point is the initial
value of the used line-frequency defined.
In the column of check boxes on the right you define which values
are
written on a file when you export analysis
results
(mean, tau, peak, etc). The same values are diplayed when you
select
View->Analysis
Results Table in the
results window.
The remaining check boxes define the following behavior:
- "Use SD for idealize": if this checkbox is NOT checked,
the
algorithm for single channel idealization
uses
a simple 50% threshold criterion. If the checkbox IS checked, the
idealization
algorithm uses the width (SD) of the peaks of the gaussians of the amplitude
histogram in order to decide to which of the peaks a given data
point
most likely corresponds.
- "Update on moving in tree": If this checkbox is checked
and
you select an ENTRY in the tree window by moving
the
arrow keys or by selecting with the mouse, the display in the trace
window
is updated. If not checked, the display is ONLY updated if you hit return
or click the right mouse button within the tree window.
- "Leak subtract": If a leak trace is stored in your data
file
together with the data trace (as is e.g. the case in HEKA-Pulse files
when
you use P/N) you can decide with this checkbox if you want to subtract
the leak or not.
- "Show Leak": If a leak trace is stored in your data file
together with the data trace (as is e.g. the case in HEKA-Pulse files
when
you use P/N) you can decide with this checkbox if you want to show it
as
a green trace or not.
- "Draw lines": if checked, traces are drawn by connecting
the individual data points by a line, otherwise they are shown as dots.
- "Segment Voltage (else: segment duration)": if checked,
the
voltage of the
the relevant segment is used
as the "x-value" for
series fits and for
exporting
analysis results (mean, tau, peak, etc). Otherwise its duration is
used as the x-value (see also
3. The relevant
segment).
- "Use 2nd trace (reading pulse-files)": if checked and if
you read a HEKA-Pulse data file, and if you have recorded TWO input
channels,
the second channel is read into Ana. Otherwise,
the
first channel is used.
- "Show Time": if checked in the
general
view the "Unix" time in seconds is displayed for series and
entries.
-"Show Stop Button": Sometimes you would like to be able to
interrupt lengthy erroneously evoked operations by a mouse click. For
example
you are fitting too many traces with too many exponentials. If this
option
is checked, a "Stop-Button" appears for several actions. It does not
work,
however, perfectly and in particluar it seems to be quite incompatible
with Windows2000.
- "Log scale in dwell time histo": if checked the
vertical
scale in the dwell time histograms is in log
scale, otherwise it's linear.
- "Dimmed overlay": if checked the traces of a
series
are drawn in grey, except the last one that remains black Otherwise all
traces are black.
-"pick events with right button": normally the right mouse
button is used in the trace-window to move the horizontal cursors. If,
however, this check-box is clicked, idealized single-channel events can
be manually edited with the right mouse button.
Parameters that are relevant for a
series
fit are accessible in a dialog that is activated by Edit->SeriesFit
Settings or Ctrl+G or by the corresponding toolbar
button.
The meaning of the various parameters is described in the
series
fit chapter.
Other settings are:
-"Settings->Hide/Show Cursors (Ctrl+H)" that toggles
on/off
the display of the
cursors in the "trace" window.
-"Settings->Hide/Show Zero line" that toggles on/off the
display
of the zero line.
-"Settings->Dwell Time Settings (Ctrl+W)" are described
in
14.3.
Dwell time analysis.
7.2. Loading/Saving
The parameter settings can be saved on a binary file (Ctrl+S,
Settings->Save
Settings, toolbar) and loaded from these files (Ctrl+L,
Settings->Load
Settings, toolbar). The last settings file that was stored is
automatically
loaded at the next execution. (The filename is saved in the ana.ini
file that is in the windows directory). You can prepare several
standard
setting files for various purposes.
7.3. Context sensitive parameters
Sometimes you might want to save a set of parameters (that partially
include
also results of analysis) in association with a specific entry or
series.
Having selected the desired series/entry and pressing X (or Settings->Save
Context Set) opens a file dialog for saving a set-file with a name
derived uniquely from the data file name and the index of the series or
entry. For reloading such a setfile you just press Y (or
Settings->Load
Context Set) and a file dialog with the same set-filename as
default
appears and hitting return will load it. You can of course also load
any
other set file thru this command.
7.4. Undoing a parameter settings change
If you have e.g. erroneously selected an unfavorable display setting,
or
you just want to return to some previous cursor
settings: just hit Ctrl+Z (or the undo button of the toolbar)
. A circular buffer keeps the last 20 display (and all other) settings
in memory. With Shift+Z you can redo the changes in the
settings.
8. Editing
commands - The internal buffer
An internal buffer keeps in memory a single entry or a whole series. It
works somewhat similar as the windows clipboard but is completely
distinct.
In particular it is defined only within a single instance of Ana
and it exists in parallel to the windows clipboard. You can use it to
transfer
data from one file to another, to delete traces, series and so on. An
important
application is that in most cases when a data-changing operation is
performed,
like e.g. permanent filtering or
deleting
of a stretch, or scaling, the original data are copied into the buffer
and the operation can be undone using the "exchange" mechanism
(see
8.1.
Undoing an erroneous operation : Exchange).
Important Note: Only the raw data and no analysis results (like
series fit, noise, fits, ...) are saved in the internal buffer. Any of
such information is lost!
In order to highlight entries in the tree (single entries or whole
sweeps) that have been altered by a data changing operation, such
entries are displayed in the tree with a differently colored icon. This
"altered" status can be explicitly reset with Changed->Remove "Changed" Status.
Saving the file will also reset this flag.
8.1. Undoing an erroneous operation : Exchange
Many operations permanently change the data (scaling, permanent
filtering,
cutting, undersampling, deleting a stretch, subtracting leak,
exchanging
data with fit, subtracting capacitance, subtracting line-frequency, and
some others) and sometimes you realize (hopefully immediately) that you
would like to undo an operation. Fortunately, Ana
makes a copy of the original data and stores it in the internal
buffer. Doing so, it overwrites (without asking) the previous
contents
of the internal buffer. So you can undo only the last operation. For
undoing
you have to select within the data tree the corresponding item (series
or entry) and then click Edit->Exchange or type Z
or
click on the double-arrow in the toolbar.
8.2. Copying/pasting/cutting/deleting/merging traces and series
With Ctrl+C (or Edit->Copy ) you copy the
currently selected
item of the data tree into the internal
buffer,
overwriting (without confirming) the current contents of the buffer.
This
applies only to single entries or single series (Note: under certain circumstances Ctrl+C or Edit->Copy, will copy
selected text in gray windows as described here).
The internal
buffer does not take more than one series, i.e. you can not copy
the
root.
Important Note: Only the raw data and no analysis results (like
series fit, noise, fits, ...) are saved in the internal
buffer. All this information is lost passing through the buffer!
With del the currently selected item is deleted without
making
a copy and thus without overriding the contents of the buffer while
with
Ctrl+X,
Edit->Cut the item is deleted and copied into the buffer. The
contents
of the buffer can be pasted before (Ctrl+V, Edit->Paste Before)
or after (Ctrl+T, Edit->Paste after) the currently selected
item.
If a series with several entries is currently stored in the buffer
these can be merged after the currently selected entry of the tree
window
by Edit->Merge entries after.
8.3. Permanent filtering
By specifying a filter frequency (larger than 0 and smaller than the
sampling
frequency) in the parameter settings
the
data are filtered only for display each time before display without
changing
the original data. You can perform a permanent, data-changing filtering
by F12 or Edit->Filter. The original data are
copied
into the
internal buffer and as long as
nothing else is copied into the internal
buffer
the filtering can be undone by the "Undoing
an erroneous operation : Exchange" mechanism.
You can also filter the whole file (i.e. all series') by Edit->Filter
whole file (Ctrl+F12) but as the internal
buffer contains at most 1 series, this command cannot be undone.
8.4. Undersampling
Undersampling by a factor of "n" means to take only every n'th point
and
to throw away the others. This reduces the number of points by a factor
of n. This can be done by Edit->Undersample or F10.
A
filtering effect can be achieved if you use Edit->Undersample
And Average.
The original data are copied into the
internal
buffer and as long as nothing else is copied into the internal
buffer the undersampling can be undone by the "Undoing
an erroneous operation : Exchange" mechanism.
You can also undersample the whole file (i.e. all series') by Edit->Undersample
whole file (Ctrl+F10) but as the internal
buffer contains at most 1 series, this command cannot be undone.
8.5. Calculating the average
With Edit->Calculate Mean (or F3) the average of
the
selected entries is calculated and placed in the internal
buffer from where it can be pasted.
8.6. Copying selected records
With Edit->Copy (Ctrl+C) the currently selected entry (if
an entry
is selected in the Tree window) or the currently
selected
series (if a series is selected in the Tree window)
is copied into the internal buffer from
where it can be pasted.
8.7. Concatenate entries
Several entries can be concatenated by this command (Edit->Concatenate)
into a single entry that is placed into the internal
buffer from where it can be pasted.
8.8. Deleting a stretch
The data stretch between the green cursors can
be deleted
by F8 or Edit->Delete piece between green cursors.
The
original trace(s) is(are) copied into the internal
buffer allwoing to undo the
deletion.
8.9. Extracting a stretch
The data stretch between the green cursors can
be
extracted (F9 or Edit->Extract piece between green
cursors
) and copied into the
internal buffer from
where it can be pasted.
8.10. Overlay envelope
Sometimes time increments or decrements are used in the pulse
protocols,
e.g. in recovery from inactivation protocols. The "relevant" or
interesting
segment then may not stay "within" the cursors for the analysis when
viewing
the traces. Data recorded by HEKA-pulse (I think not pclamp-data) have
a variable number of samples for each record in such cases. This allows
the following trick: applying the Even more analysis->overlay
envelope
command Ana adds zeros to the beginning of each
trace
such that all traces will have the same number of samples resulting in
a perfect superposition of the segments after the variable-length
period.
8.11. Swapping data and fit
If a trace fit has been performed, that is
shown in blue, the blue, fitted, stretch can be swapped with the
corresponding
data stretch by the command
Even more analysis->Swap data-fit.
8.12. Reverse time
With this command (shortcut = Shift+T) you can reverse the
time
of one or more entries. This could be useful to test if your
(single-channel)
analysis depends on the direction of time or, more profanely, you can
use
this to extrapolate a fit not to a previous time point but to later
times.
If more than one entry is subjected to the time-reversal you can chose
to reverse also the order of the entries to get a complete time
reversal,
or not. In the former case the pulse-protocol (if varying voltages or
times
are involved) WILL NOT BE VALID AFTER THE REVERSAL!
8.13. Paste as fit
With the command Edit->Paste as fit (shortcut = F5)
the
internal
buffer content is pasted as a trace
fitting
(i.e. a blue line) overriding any fit present, starting at the
beginning
of the trace! With Edit->Paste as fit between green cursors (shortcut
= Ctrl+F5) the
internal buffer
content is pasted as a fit only between the green cursors.
9. Trace fitting
The data traces can be fitted by a variety of functions, in particular
exponential functions with various constraints, polynomials, and, using
an equation-parser, any user-defined function .
The following general rules apply:
The fit is displayed on top of the data in blue color.
Fitting is performed using the Simplex algorithm (except for the
polynomials
that are calculated directly). The number of iterations and the
stepsize
are defined in the Series Fit parameters
dialog
.
The fit is applied to a single entry if a single entry is selected
in
the tree window or to a group of entries within a
series
if a series is selected in the tree window.
The trace is fitted between the two green cursors,
i.e. the parameters of the fitted function depend on the data values
between
the green cursors.
If the blue cursor is located to the left of the left green cursor,
the fit is extrapolated up to the blue cursor.
This
is for example used to estimate the initial current value after a
voltage
jump that is disturbed by a capacitive artifact.
A single entry can "contain" only a single fit. A new trace fit
overrides
the old one. This implies that all results
values that depend on the fit are also overwritten (e.g. the "initial
value").
The fits are saved on the file if it is
saved.
If entries are copied into the internal
buffer , the fit is not copied.
The data trace and the fit can be "exchanged".
This does, however, not lead to a recalculation of the initial value or
other values that depend on the parameters of the fit.
The entries of the internal buffer can
be "pasted as fit", i.e. any old fit trace is
overwritten and the pasted trace is shown in blue. Pasting starts from
the first data point (i.e. the position of the cursors have no
influence).
This is used mainly for display purposes. It does not lead to a
recalculation
of the initial value or other values that depend on the parameters of
the
fit.
9.1. Exponential fits
In all exponential fits traces are fitted by functions of the form
a0 + a1×exp(-t/tau1) + a2×exp(-t/tau2) + ... +
an×exp(-t/taun)
n, the number of exponentials, must be at least 1. It is determined
in the general parameter settings dialog
as "number of components". The time point 0 is determined by the left
green
cursor
if the blue cursor is located to the right of
it
and otherwise by the position of the blue
cursor
(used for backextrapolation, see 4. Cursors).
In
a simple full free exponential fit (Ctrl+E, toolbar) all
parameters
(ai, taui) are freely fitted, while several specially constrained
exponential
fits are described below. Of particular importance is the common
exp fit which is used to fit the same time constant(s) for all
entries
in a constant-voltage tail protocol to extract the steady state popen
from
tail currents. The parameters (see "initial value (from fit)", "fast
tau",
"slow tau", "very slow tau", "coefficients", in the general
parameter settings dialog) of exponential fits can be exported
on
a file or displayed as a table and
selected
with the mouse. The initial values for the time constants are guessed
based
on the time interval. Sometimes these guesses are not appropriate and
manual
intervention is necessary. In this case you can try to play with the
stepsize
and the number of iterations (see tracefitting).
If this does not help you can manually enter starting values by hitting
"i" on the keyboard while the fit is running (this works only if the
checkbox
allow
keyboard input in fit (see Series Fit))
in
the series fit dialog is checked). If also this does not help you can
try
a user-defined function where the initial
values
have to be manually set. Note that the coefficients, ai, are not
dependent
on any initial value assumption as they are optimally calculated for a
given set of time-constants.
9.1.1. Simple exponential fit
In a simple exponential fit all coefficients and time constants are
freely
fitted. See '9.1. Exponential Fits' for
a
general description.
9.1.2. Fit several traces simultaneously with the same
time-constants for
all
In this fit several entries are fitted simultaneously with the SAME
time
constant(s) for all entries. The coefficients are calculated
individually
for all entries. A frequently used protocol to estimate the
steady-state
voltage-dependence is to apply a pulse to a fixed voltage after
prepulses
to varying voltages. The initial current at the constant "tail" pulse
is
proportional to the open-probability at the end of the prepulse. The
"common
exp" fit is generally used to estimate the initial value by exponential
fitting and backextrapolation. It yields also a good estimate of the
time
constant(s) at the fixed tail voltage.
9.1.3. Other exponentials
Somewhat differently treated from the normal sum of exponentials is the
product
of two exponentials (Analysis->Special Exps->fit product of
two exps)
because the coefficients have to be fitted too. A variant of this is Analysis->Special
Exps-> product of two exps, fix one in which one of the time
constants
is fixed.
In the normal "sum-of-exponentials fit", there is also the option to
keep one time constant fixed, either for each trace singly (Analysis->Special
Exps->fit exps, fix one) or keeping the same taus for all
traces (Analysis->Special
Exps->fit common exps, fix one; see 9.1.2.
Fit several traces simultaneously with the same time-constants for all).
Another option is to constrain the coefficients in a way to keep the
initial derivative equal to zero:
dY/dt( t=0) = 0
Try Analysis->Special Exps->fit exps with DI(0)=0 (Ctrl+D)
and Analysis->Special Exps->fit common exps with DI(0)=0.
9.1.4. Envelope
Not yet documented. But see: 8.10. Overlay
envelope.
9.2. Polynomial
The "number of components" in the parameter
settings dialog determines the number of coefficients of the
polynomial
to be fitted, i.e. 1 corresponds to a horizontal line, 2 is a straight
line, 3 is a parabola, etc. The coefficients can be visualized in the Results
window from where they can be copied and pasted.
9.3. User defined
With More Analysis->Fit trace with user function or Ctrl+U
the fit of the trace(s) by an arbitrary user-defined function is
invoked.
The function is defined in the series-fit
dialog.
The time-variable can be written as "t" or as "v".
The
parameters of such a fit are written as a[0], a[1],
etc.
and the initial values are defined in the dialog invoked by clicking Settings->Edit
Params or typing E. Here you also specify for each
parameter
if you want it to be adjusted (checking the corresponding "DoFit" box)
or if it should remain fixed.
As an example you can compare the intrinsic exponential fit with the
following user defined function:
(a[0]+a[1]*exp(-t/a[2]))*1e-12
The factor 1e-12 here is just for convenience to keep the parameters
a[0] and a[1] in pA. Time is always in SECONDS!
See 18.3. Syntactic rules for the user-defined
function
for the syntactic rules of the user-function.
10. Series Fit
Series fit means that you display and fit a particular value (e.g. the
mean calculated between the 2 green cursors)
for
all the entries of a series. The x-value can be either
- the voltage of the
relevant segment
(if the checkbox "Segment Voltage (else: segment duration)" in
the
parameter settings dialog is
checked).
- the duration of the
relevant segment
(if the checkbox "Segment Voltage (else: segment duration)" in
the
parameter settings dialog is NOT
checked).
- the entry-index (starting with 0 for the first entry) defined if
you
set the
relevant segment to -1
- the absolute time of acquisition of the entry (starting with 0 for
the first entry) defined if you set the relevant
segment to -2
The type of function to fit is selected in the Series Fit
settings
dialog
(Ctrl+G) . The radio buttons on the top left of the dialog
decide
what function you want to fit. (see below for a precise definition). In
the column of radio buttons on the right you select which value you
want
to fit. The other fields mean:
- # components in poly fit: The number of coefficients for a
Polynomial
fit.
- Maxiter: the number of iterations in the fit.
- Stepsize: the stepsize for the simplex search in the fit.
- User function: here the user function is defined. It is
used
either by the user defined trace fit or by
the
User
defined series fit.
- Bin width ampl histo (in pA) : The bin-width for the Amplitude
Histogram
- Bin width dwellt. histo (in ms) : The bin-width for the Dwell
time analysis
- min dwelltime (ms): The minimal dwell-time for the Dwell
time analysis
- Invert Y-data : if checked the values to be analyzed are
multiplied
by -1.
- Equidistant gaussians : if checked the peaks of the
gaussians
are equidistant in the Amplitude
Histogram
- Allow keyboard input in fit: only if checked has keyboard
input
an influence during trace- and series-fit. With "s" the fit is
interrupted.
With "i" new initial values of the various parameters can be entered
during
fitting.
The data values and the fit are shown in the upper window.
Clicking
with the right mouse button on the individual data points you can
display
the corresponding x and y value.
10.1. Boltzmann
In a Boltzmann-fit the following function is fitted:
offset + (Max-offset)/(1+exp( (v-V1/2)/k ))
with the four parameters, offset, Max, V1/2,
and k. If you want to fit a different kind of function (e.g.
fix
the offset to 0) you have to use the User
defined series fit.
10.2. Polynomial
A polynomial is fitted with the number of coefficients, Ncoeff, as
described
above (see 10. Series Fit). With NCoeff=1 the
mean is calculated (and displayed). With NCoeff=2 a straight line is
fitted
and assuming that the SeriesFit is an IV also the extrapolated reversal
potential is calculated and displayed. With NCoeff=3 a parabola is
fitted
and assuming that the SeriesFit is an IV also the two possible
extrapolated
reversal potentials are calculated and displayed. For Ncoeff>3 only
the
coefficients are displayed.
10.3. User defined function
The function is defined in the series-fit
dialog.
The independent variable can be written as "v" or as "t".
The parameters of such a fit are written as a[0], a[1],
etc. and the initial values are defined in the dialog invoked by
clicking
Settings->Edit
Params or typing E. Here you also specify for each
parameter
if you want it to be adjusted (checking the corresponding "DoFit" box)
or if it should remain fixed.
As an example you can compare the intrinsic Boltzmann
fit with the following user defined function:
a[0]*(a[1]+(1-a[1])/(1 + exp((v-a[2])/a[3]))*1e-12
The factor 1e-12 is to keep the parameter a[0] in pA. You can fix
e.g.
the "offset" to zero by entering a[1]=0 in the dialog and not fitting
the
value.
See 18.3. Syntactic rules for the user-defined
function
for the syntactic rules of the user-function.
11. Leak, capacity, and line frequency (50/60 Hz) subtraction
Before these various subtractions, the original data are copied to the
internal
buffer and the operations can be undone using the "exchange"
mechanism (see
8.1. Undoing an erroneous operation
: Exchange).
11.1. Baseline subtraction
With Leak Subtraction->subtract baseline or Ctrl+B
the
data between the two red cursors is averaged
and
subtracted from the trace(s).
11.2. Leak subtraction
Ana offers 2 possibilities to perform offline
leak
subtraction. The first is two subtract a scaled series of leak records,
the second is based on a baseline segment ...
no help yet, sorry.
11.2.1. Subtract a scaled series
no help yet, sorry.
11.2.2. Subtract using a baseline segment where channels are
supposed to
be closed
no help yet, sorry.
11.3. Capacity correction
Suppose one segment of the stimulation pulse goes to a voltage where no
current flows because the driving force is 0 (i.e. a step to the
reversal
potential). You can use this segment to subtract the capacity from
other
segments as in the following example. Suppose your protocol is like
this:
segment 0: VHold, 10 ms
segment 1: VTest1, 10 ms
segment 2: VTest2, 5 ms
segment 3: VHOLD=EREV, 10 ms
You can subtract the capacitive artefact from segments 1 and 2
(if
VTest1 and VTest2 are different from EREV). Let's do it for segment 1.
Select the sweep (or series) where you want to subtract the capacity.
Put
the "relevant segment" to 1. Select "View->Put green cursors on
segment"
(Ctrl+-) to put the green cursors on segment 1. Select "View->Put
red cursors on segment" (Ctrl+<) to put the red
cursors
on segment 3!
Select "Leak Subtraction->Separated segments capacity correction"
(Ctrl+F11) to subtract the capacity. Select "3" as the "leak"
segment.
The original data are copied into the
internal
buffer and as long as nothing else is copied into the internal
buffer the subtraction can be undone by the "Undoing
an erroneous operation : Exchange" mechanism.
You can of course also manually put the cursors to fit exactly the
start of the segment if there is an incongruence with the stimulation
protocol.
11.4. line frequency (50/60 Hz) subtraction
no help yet, sorry.
11.4.1 Automatic line frequency (50/60 Hz) subtraction
no help yet, sorry.
11.4.2 Manual line frequency (50/60 Hz) subtraction
no help yet, sorry.
12. Scaling traces
12.1 Scaling by a factor and add a constant
With the command MoreAnalysis->ScaleTrace (or via the
toolbar)
the selected entries are transformed by
x -> m*x + b
with a factor m and a constant b. Note that b
must be given in units of Ampere. The original data are copied into the
internal
buffer and as long as nothing else is copied into the internal
buffer the scaling can be undone by the "Undoing
an erroneous operation : Exchange" mechanism.
12.2 Scaling traces of a series to have a common value in a certain
segment
With the command MoreAnalysis->ScaleTails (or hitting "J")
the entries are scaled in a way that the mean of the stretch delimited
by the green cursors is the same for all and
corresponds
to one that can be selected. This is e.g. useful if you are recording
an
instantaneous IV and there is run-down and you want to scale the traces
to have the same prepulse amplitude. The original data are copied into
the
internal buffer and as long as nothing
else is copied into the internal buffer
the scaling can be undone by the "Undoing
an
erroneous operation : Exchange" mechanism.
13. Noise analysis (non-stationary!)
Non-stationary noise analysis serves to estimate the single-channel
conductance
and the number of channels. For a theoretical background please refer
to
1. Sigworth , 1980, The variance of sodium current fluctuations at
the
node of Ranvier. J Physiol. 307:97-129.
2. Heinemann and Conti , 1992, Nonstationary noise analysis and
application
to patch clamp recordings. Meth. Enzymol. 207:131-48.
Briefly, a fixed pulse during which the channels undergo a
time-dependent
relaxation is applied repeatedly, e.g. 50 times. The analysis is
divided
into three steps:
1. Selection of the time stretch and the background stretch to be used
(if any)
2. Selection of records
3. Variance-mean analysis with a parabolic fit
13.1. Selection of records and cursor settings
The stretch of the record that is used for the noise analysis is
defined
by the green cursors. If you want to subtract a
baseline variance you have to position the red cursors
over the corresponding stretch.
It is often advisable to eliminate "bad" records that contain e.g.
disturbances
or unwanted electrical activity from the set of recorded traces to be
used
for the noise analysis. One possibility is to go manually through the
records
and throw out those that appear to be bad.
Ana offers
also a somewhat more objective possibility to detect bad records. As a
criterion Ana identifies a record as
potentially bad
if it deviates at a single data point from the crude mean by a certain
number of S.D.'s. This factor is defined in the general
parameter settings dialog.
With Analysis->Noise etc->NoiseAnalysis or Ctrl+N
the selection of the records is started. First you have to select the
range
of records for which the noise-analysis is to be performed. If you want
to use all records, no further selection is necessary. Also, if you
have
done previously a selection (that is saved on a "lst"-file; see below)
that you want to use, no further selection is necessary; just confirm
the
corresponding questions. If you want to select the records manually,
follow
the instructions: In the trace window the single
records are now displayed overlayed with the cumulative mean of the
selected
traces (apart from the first trace, that is shown without mean). If a
trace
has been identified as potentially bad by
Ana, this
will be written on top. The selection has now to be done with the
keyboard:
- to accept a trace as good: hit "Y" (yes)
- to not accept a trace: hit "N" (no)
- to stop the selection process and proceed the analysis with the
traces
selected sofar: hit "S" (stop)
- to completely interrupt the noise-analysis: hit "Q" (quit)
- to go back one record: hit "B" (back)
- to rush, i.e. to accept automatically all good records and to ask
explicitely only for the potentially "bad" records: hit "R" (rush)
After finishing the selection it is written on a file with the same
name as the data-file but with the extension ".lst". If such a file
exists
already you are asked if you want to overwrite the old selection. Thus,
the selection of the good records is saved and you don't have to redo
it.
You just answer "yes" to the question "Use old selection(lst)?", the
next
time.
Because the selection file is simply a list based on the indexes of
the traces it is not any more applicable if you change the data file by
deleting or adding records. To avoid such a possible error you can also
"extract" the selected records from a series by Edit->Extract
records,
that will put only the selected records as a series into the internal
buffer that you can paste for example in an empty file.
After the selection - using either all records, or using an old
selection,
or by going through the selection process as described above - the user
is asked for "the number of segments". This has the following meaning:
for the variance-mean plot one often does not want to display all data
points but to have a more condensed representation. For this purpose,
the
current range of the calculated mean (within the selected stretch
(green
cursors))
is horizontally sliced into segments (this is the "number of
segments").
This cuts the current trace into several segments of equal vertical
extension
but possibly variable length. Rapidly changing parts are thus shorter,
while a long plateau could be fit into a few segments. Within each
segment
of the mean current trace the current and the corresponding variance
values
are averaged and yield one point in the variance-mean plot (see also
Sigworth
, 1980, J Physiol. 307:97-129 and Heinemann and Conti , 1992, Meth.
Enzymol.
207:131-48). If you don't want to do this kind of averaging you can
enter
a very high number as the number of segments (e.g. larger than the
number
of points between the green cursors).
The result of the noise analysis so far is that the mean current and
the variance has been calculated (and stored; you can display them as
described
in 2.2. Traces window) for the whole trace.
Furthermore,
for the part between the green
cursors the current
and variance values that come from the slicing procedure described
above,
are stored in an extra array that can be further analyzed by the
variance-mean
parabolic fit as described below. Also the (average) variance between
the
red cursors has been calculated as background
variance,
that can be subtracted (or not) for the variance-mean analysis.
A remark regarding the sign of the mean currents: Ana
automatically converts a negative current range (between the green cursors)
into a positive range by multiplying with -1!
13.2. Variance-mean analysis
By Analysis->Noise etc->Var Mean, or Ctrl+J
the variance-mean
parabolic fit dialog is invoked. This does not work if you haven't
previously
calculated the mean and variance as described above (if you have loaded
a data file that was previously saved with Ana
after
having calculated the variance, the variance is also loaded; you don't
have to redo the calculation; see 18.2. The
internal
data-format of ana: what is saved on a file?). In the dialog you
specify
the initial values for the parameters, the number of iterations, which
parameters are to be fit and how the background variance and the leak
has
to be treated. The fit is according to the following formula
sigma2 = sigma02 +
i*(1+o^2)*(I-leak)
- (I-leak)2/N
,
where sigma02 is the background
variance,
leak
is the leak current, i is the single channel current,
N
the number of channels, o is the "fraction open channel noise"
(normally
= 0; see below), and I the independent variable, the mean
current.
It is, of course, not advisable to have both, leak and background
variance,
as free parameters. Instead it is better to have as most as possible
independent
measurements for these two parameters, and to keep them fixed. After
clicking
OK
in the dialog, the parabolic fit is performed and the results are
displayed.
You can export the results of the variance analysis by File->Export
Analysis->Export Var-Mean. The results are written on a plain
white-spaced
ASCII file as columns. If you answer "YES" to the "Write trace?"
question,
the first three columns of the file contain the time, mean current (in
pA), and mean variance values (in pA^2) of the whole trace. Note that
the
background variance has NOT been subtracted yet from the variance
values.
The next three columns contain: mean current of the segments, mean
variance
of the segments, and standard deviation of the variance. The background
variance is written in the last (single-valued) column.
Note on the parameter "o" (fraction of open channel noise): The
"profane" user should just ignore this value (setting it to 0). It is
not
a real parameter to fit as it is absolutely not independent from the
parameter
i. However, for the calculation of the maximal open probability it is
sometimes
useful to have an idea about how an "open channel noise" would
influence
the estimate of pmax. The idea is that with all channels open the
variance
is not zero, but equal to N*sigmaopen2, where
sigmaopen2
is the variance of an open channel that is parameterized as sigmaopen2=(o*i)2,
hence the meaning of the parameter o. If not all channels are open, the
open channel noise is assumed to behave as N*p*sigmaopen2,
and putting this into the normal variance-mean-equation results in the
above formula.
14. Single channel analysis
Two basic features are offered for single channel analysis: amplitude
histogram analysis and dwell-time analysis.
The dwell-time analysis can only be done after having constructed and
fitted
the amplitude histogram by a sum of Gaussians, because in this way the
levels are defined that are used for the automatic
idealization. Note that the idealization is COMPLETELY AUTOMATIC;
it
is not possible to change the single transitions. This possibility
might
be incorporated in future versions.
14.1. Amplitude Histogram and Gaussian Fit
To construct the amplitude histogram select Analysis->Construct
Ampl.
histogram (or Shift+H or via the toolbar). A dialog
appears
where you select
- if you want to restrict the analysis to the region between the green
cursors
- the first and last entry to analyze
- the bin width used for the histogram
- the current-range
Then you fit the amplitude histogram with a sum of Gaussians
by
selecting Analysis->Fit Ampl. histogram (or Ctrl+M
or
via the toolbar). A dialog appears where you select
- the number of gaussians (identical with the "number of components"
of the general settings)
- if the gaussians shall be equidistant
- how the initial parameters of the Gaussians shall be chosen
- if the fitted parameters should be copied in the "parameter array"
after the fit
- the stepsize and the number of iterations of the fit
- if keyboard input is allowed during the fit (this option turns on
e.g. the possibility to interrupt the fit by hitting "s" (stop);
however
as the keyboard input is checked asynchronously any "s" typed during
the
fit, also if in another program, like e.g. a text-writing program, will
interrupt the fit and you might want to disable this feature such that
you can write during a lengthy fit).
The program will likely NOT find automatically the correct
parameters
for a multi-Gaussian fit but they have to be selected manually in a
simple
graphical fashion described below, requiring thus at least two rounds
of
the Gaussian fit. The first time you better select "Automatic
re-calculation"
for the start params that leads to a practically random placement
of the mid-points of the Gaussians. Each Gaussian component has a
vertical
green line at the midpoint, a horizontal green line at the peak value
and
circles at the ends of the horizontal line indicating the sigma-value
of
the Gaussian. The midpoint (vertical lines) can be moved with the left
mouse button. The amplitude (vertical line) of the Gaussians can be
moved
with the right mouse button and the sigma value can be changed by
moving
one of the circles with the left mouse button. You should thus adjust
manually
all Gaussians in an approximate manner and then select again Analysis->Fit
Ampl. histogram (or Ctrl+M or via the toolbar) but now
selecting
the option "Use last values" for the start parameters. Also the step
size
should not be too big ( a value of 0.1 is normally ok together with
about
1000-10000 iterations, depending of course on the number of Gaussians
and
how well they are defined by the data).
If you select the option "equidistant means", the LEFTMOST
midpoint
and the next one are used for determining the position of the others!
From the Parameters of the Gaussians the relative areas under each
component
are calculated (in %) and displayed together with the other parameters
in the grey selectable and copyable text box on the right.
Clicking
on the button "Copy params to array" copies the parameters into the
data
array that is reachable by hitting "E" (or via the menu: Settings->EditParams).
This array is part of the settings.
It
can be used here to define precise starting values and/or to save the
gaussian
parameters in a setting file from where they can be reloaded as
starting
values as described above.
Once the Gaussians are fitted you can proceed with the Idealization.
14.2. Idealization
Once the amplitude histograms has been fitted
with
a sum of Gaussians, the midpoints of the Gaussians are used as the
levels for the idealized current traces. Two different kinds of citeria
can be applied for the idealization:
- 1. a simple 50% threshold criterion or
- 2. a criterion based on the width (SD) of the respective Gaussians
(as explained below).
Which of these criteria is used is selected in the General
Settings dialog in the check box "Use SD for idealize". If checked,
criterion 2, otherwise criterion 1 is applied. Criterion 2 works as
follows:
Suppose there are n levels, mi is midpoint of level i, vari is the
SD^2 of level i, and y is a data point that has to be assigned to a
level.
For each level the quantity pi=exp( -(mi-y)^2/ 2 / vari) is calculated
and the level with the largest pi is chosen as the most likely one.
(Actually the pi's are calculated as (mi-y)^2/vari and the smallest
pi is selected. The result is the same).
Idealization is dependent on a further parameter that is asked for
when "Analysis->Idealize" (or Ctrl+I) is
selected: the
minimal number of data points that an acceptable event must be long.
The idealized trace is shown in blue and represents as such a "fit"
of the data trace, implying that any other fit, like fitting an
exponential
will override and replace the idealization. Saving a data file in this
status will save also the idealization, as any other fit.
14.3. Manual editing of the idealized records
It may happen that some events are not acceptable (e.g. too
short)
after the idealization. These can be eliminated in the following way:
first,
manually editing has to be enabled by a check-box-flag in the Settings
dialog: make sure that the check box "pick events with right
button"
is checked in the settings dialog. This makes sure that the right
button
in the trace-window does not move the horizontal cursors but picks the
events. Then place the mouse over the event that you want to change and
press the right button. A popup-menu appears with various choices. The
last action (elimination) can be undone if, and only if, the
corresponding
stretch is at least partially visible on the screen.
14.4. Dwell time analysis
Like the amplitude-histogram, the dwell times are delt with in 2 steps:
1st construction of dwell time histograms, then 2nd. fit by a sum of
exponentials.
Several important general features apply, however, to the dwell time
analysis:
- The fit by the sum of exponentials is completely independent of
the
representation in a histogram (bin width etc.). All events of a given
level
are explicitely taken into account (larger than a minimum dwell time to
be specified) and the fit is done using a Maximum Likelihood criterion
as detailed below.
- The dwell time histograms are cumulative histograms with a linear
time scale. The vertical scale can be either linear or log dependent on
the setting in the general parameters
dialog(also
selectable in the fitting dialog described below).
Selecting Analysis->Construct Dwell Times (or just
hitting
O,
or toolbar) the dwell time histograms for all levels are constructed
and
displayed. Here, the only important parameter is the bin width of the
histograms,
to be selected in the series fit settings.
Selecting Analysis->Fit Dwell Times (or just hitting
Q,
or toolbar) the dwell times of the specified level are fitted by the
sum
of exponentials as specified in the dialog that appears. Here you
select
- the number of exponentials
- start values for each time constant and coefficient to be fitted
- if you want to fit or otherwise fix the time constant and/or
coefficient
- some other parameters
Note that the LAST COEFFICIENT WILL ALWAYS BE VARIABLE because the
sum
of the coefficients is constrained to be 1, such that the number of
effective
parameters is reduced by 1, excluding the last coefficient as a
fittable
parameter.
The fit is done according to the Max.Likelihood criterion i.e. for
each
event, e, of duration te, the likelihood is calculated as
Le= sum over i (a[i]*exp(-te/tau[i])) / LMINMAX
where LMINMAX is calculated as
LMINMAX=sum(a[i]*(exp(-MinDwell/tau[i]) - exp(-MaxDwell/tau[i]))
and is the probability that an event has a duration between MinDwell
and MaxDwell. The parameter MinDwell has to be specified in the above
mentioned
dialog. The parameter MaxDwell is determined from the shortest length
of
the records used for the construction of the dwell time histogram.
14.5. Binomial Peak analysis
Sometimes, for example if several channels are in a patch, one would
like to perform a binomial analysis of the areas of the amplitude
histogram. One reason is to determine the open-probability assuming the
presence of N independent channels. Or, the independence of the
channels shall be tested. For these purposes, the "Binomial Peak Fit"
has been implemented. It can be assessed via "Analysis->Fit Binomial
Distribution" that invokes a start-dialog. Data can be entered in the
white window. The format is "n A F", for each data point, where n is
the associated current level, A the experimental probability (in %) and
F is the fitted probability. The latter can be set to any value for
input and can be used to read out the fitted value after the fit.
Let's take an example. Assume two channels, each with popen=p=0.5. The
"ideal" experimental data would be
0 25 0
1 50 0
2 25 0
I.e. Prob=0.25 to have no channel open, 1/2 that one channel is open
and 0.25 that both are open.
You can enter the data either manually, or, better, paste them from
another program.
NOTE THAT TO MAKE A NEW LINE IN THE DATA INPUT WINDOW YOU TO PRESS
CTRL+ENTER!
If (and only if) you have previously fitted an amplitude histogram, the
corresponding area values are shown initially as experimental values in
the input window. You don't need to use these, however. By clicking on
the "Copy Data from amplitude histogram" button, the data from the
amplitude histogram (if that is present) will be copied into the data
window and overwrite any other data there.
Three options are offered for fit.
1. The simple binomial fit. This is the usual case. Each level, k, has
probability
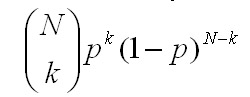
Note that the parameter N can be selected as a free parameter.
However, for numerical reasons, the Simplex algorithm might not find
the best value. Try to do the fit from several starting values for N
(and reasonable values for p=p1) and check the resulting SD value shown
after the fit. The best fit has the lowest SD value.
For this simple binomial fit the parameters p2 and N2 are irrelevant.
2. The second case assumes two "modes" of the channel, one mode with
p=p1 and another mode with p=p2. N is the total number of
channels, while N2 is the number of channels in mode 2. This
N1=N-N2 is the number of channels in mode 1.
Again, the N's can be chosen as free parameters but the Simplex
algorithm has difficulties finding the true minimum!!
The formula for calculating the probability for each level, k, in this
case is
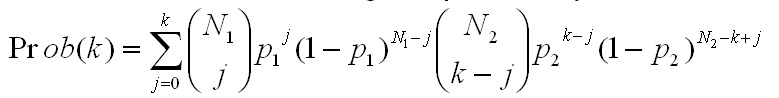
with the definition that (n over j) = 0 for j>n.
3. The third option assumes a double-barreled channel with a common
gate with popen=ps=p1 and a protopore gate with popen=pf=p1. The two
gates are assumed to be independent.
After performing the fit, experimental values are shown as bars, fitted
values are shown as symbols.
15. Spectral analysis
no help yet, sorry.
16. Exporting data
16.1. Export raw data
Raw data can be exported in ASCII format. The format is in columns with
column 1 containing the time, column 2 containing the data of the first
entry, column 3 the data of the 2nd entry, ... The columns are
separated
by "white space". You can chose to export either all data or only
between
the green cursors. The default time/current units are seconds/Amperes
but
there is also the possibility to scale these to export either in ms/pA
or
ms/microA. The Biopatch ASCII format can be read by an analysis
program
of Biologic. The other options (autocovariance, average between
cursors)
are quite special and not further documented at this time.
Since Jan. 2006, you can export data also using cut and paste. Selecting View->Numbers the upper
right window displays the selected traces as columns of numbers that
can be copied into the Windows clipboard (right mouse button!) and
pasted into Excel ot other programs. The data format of the numbers
(i.e. time in seconds or milliseconds, current in A or pA or microA, or
raw data) is defined in the Settings->Export/Table
scalings dialog.
16.2. Export analysis results (mean, tau, peak, etc)
no help yet, sorry.
16.3. Export noise analysis
You can export the results of the
noise analysis
by File->Export Analysis->Export Var-Mean. The results
are written
on a plain white-spaced ASCII file as columns. If you answer "YES" to
the
"Write trace?" question, the first three columns of the file contain
the
time, mean current (in pA), and mean variance values (in pA^2) of the
whole
trace. Note that the background variance has NOT been subtracted yet
from
the variance values. The next three columns contain: mean current of
the
segments, mean variance of the segments, and standard deviation of the
variance. The background variance is written in the last
(single-valued)
column.
16.4. Export spectrum
no help yet, sorry.
16.5. Export amplitude histogram
You can export the results of the
amplitude
histograms analysis by File->Export Analysis->Amplitude
Histogram.
The results are written on a plain ASCII file as columns with "white
space"
separating the columns. Column 1 contains the current bins. Column 2
contains
the data. Column 3 contains the Gaussian fit. Column 4 contains the
parameters
of the Gaussian fit in the order: mean, sigma, amplitude, area (in %).
16.5. Export dwell time histogram
You can export the results of the
dwell
time histogram by File->Export
Analysis->ExportDwellTimeHisto.
The results are written on a plain ASCII file as columns with "white
space"
separating the columns. The data for the various levels are written on
top of each other, separated by a blank line. Column 1 contains the
time
bins. Column 2 contains the data and column 3 the fit (if present) with
the sum of exponentials.
17. Misc.
17.1. Changing the pulse protocol ("Stimwave")
no help yet, sorry.
18. Appendix
18.1. Supported data file types
Below is a list of files that can be read by Ana.
For Axon binary files (abf-files) it was sometimes difficult to get the
correct gain-settings. For this reason, the "abf-gain-factor" has been
introduced in the Parameter Settings
that
allows the user to multiply the default-read gain by a factor in order
to get the correct amplitude, but this should not be necessary anymore.
For all files you can a posteriori change the gain in the Edit->SeriesParameters
for each series individually (maybe tedious).
- HEKA Pulse Version 7: Select the ".dat" file (the pul and pgf must
of course be present)
- HEKA Pulse Version 8: Select the ".dat" file (the pul and pgf must
of course be present)
- Axon abf
- pclamp Clampex 5.5.
- pclamp Fetchex 5.5.
- Biologic DTR file (digital tape recorder)
- Atari Pulsed (Affolter), dataversion 200
- Pulse++ of Uli Rexhausen (only version 1.2)
- GePulse: data files generated by the free acquistion program written
by M. Pusch (for National Instruments PCI-6035E and PCI-6052 and
now also many other NI interfaces and
Instrutech ITC-16 and ITC-18 interfaces). Download
here.
If you are interested in reading YOUR favorite data file,
send
me a documentation and an example and I will try my best.
18.2. The internal data-format of ana: what is saved on a file?
All the original data and the performed analysis is saved on the file,
but not the
parameter settings. For a
rough description of the file format, see below. If you are interested
in a precise description, see
Data-Format (detailed).
If you select File->Save As Ana
proposes
as default a name that is distinct from the original data file name in
order to avoid that you overwrite it. The "standard" but not required
extension
is "EAT".
The data file (version 6.0) is written by the following procedure:
The
2 procedures used are:
WriteBytes(CFile * fp, UINT pos /* file positions */, UINT
NumberOfBytes,
BYTE * variable); and
"WriBy" is a macro that writes a variable in binary format using the
size of the type as the number of bytes.
WriteBytes(fp, pos, 64, Version);
WriteBytes(fp, pos, 12, MainUnits);
WriBy(ADCConversion)
WriBy(NumberOfChannels)
WriBy(Temperature)
WriteBytes(fp, pos, 256, buff);
if (versionnumber>4){
RootText.WriteOnFile(pos, fp); // Writes the
text assigned to the root. The size depends on the string-length (if
any).
}
WriBy(NEntries)
WriBy(NSeries)
for (__int32 i=0; i<NEntries; i++){
WriBy(entries[i].NData)
WriBy(entries[i].FilePos)
WriBy(entries[i].P4Present)
WriBy(entries[i].Temperature)
WriBy(entries[i].Time)
WriBy(entries[i].IsBold)
WriBy(entries[i].s1) // unused variables
WriBy(entries[i].s2)
WriBy(entries[i].s3)
WriBy(entries[i].s4)
if (VersionNumber>4){
entries[i].EntryText.WriteOnFile(pos, fp);
}
} // end for entries
for ( i=0; i<NSeries; i++){
WriBy(series[i].VHold)
WriBy(series[i].Filter)
WriBy(series[i].StimInterval)
WriBy(series[i].SampleTime)
WriBy(series[i].Gain)
WriteBytes(fp, pos, SERIESCOMMENTSIZE, series[i].Comment);
WriBy(series[i].firstrec)
WriBy(series[i].NPulses)
WriBy(series[i].NumAveraged)
WriBy(series[i].P4Present)
WriBy(series[i].P4VHold)
if (VersionNumber>4){
series[i].SeriesText.WriteOnFile(pos, fp);
}
WriBy(series[i].NSegs)
for (int k=0; k<series[i].NSegs; k++){ //MaxSegs;
k++) {
WriBy(series[i].Pulses[k].A)
WriBy(series[i].Pulses[k].T)
WriBy(series[i].Pulses[k].SV)
WriBy(series[i].Pulses[k].ST)
WriBy(series[i].Pulses[k].FV)
WriBy(series[i].Pulses[k].FT)
WriBy(series[i].Pulses[k].SK)
WriBy(series[i].Pulses[k].s1)
WriBy(series[i].Pulses[k].s2)
WriBy(series[i].Pulses[k].s3)
WriBy(series[i].Pulses[k].s4)
}
WriBy(series[i].IsBold)
WriBy(series[i].s1)
WriBy(series[i].s2)
WriBy(series[i].s3)
WriBy(series[i].s4)
WriteNoise(fp, pos, i);
WriteSpectrum(fp, pos, i);
series[i].Histo.Write(fp, pos);
series[i].VM.Write(fp, pos);
series[i].Lore.Write(fp, pos);
series[i].SF.Write(fp, pos);
series[i].DwellTimes.Write(fp, pos);
for (int kk=0; kk<19; kk++){
BOOL dummy=FALSE;
WriBy(dummy)
}
}
for (i=0; i<NEntries; i++){
WriteBytes(fp, pos, entries[i].NData*2, entries[i].IntData);
if (entries[i].P4Present) {
WriteBytes(fp, pos, entries[i].NData*2,entries[i].P4Data);
}
if (versionnumber>2){ // Write Fit and Results
WriteFit(fp, pos, i);
results[i].Write(fp, pos);
for (int kk=0; kk<20; kk++){
BOOL dummy=FALSE;
WriBy(dummy)
}
}
}
18.3. Syntactic rules for the user-defined function
The independent variable can be written as "v" or as "t".
Parameters are a[0], a[1], etc. The index, i, in a[i] must be a
constant
number (i.e. not an expression).
Arbitrary nested expressions (with parenthesis) are allowed.
Spaces are ignored. No distinction between upper- and lowercase is
made.
Allowed functions are:
a+b
a-b
a*b
a/b
a^b
exp(a)
exp10(a)=10^a
inv(a)=1/a
sqrt(a)=square root
log(a)=natural log
log10(a)=log base 10
sin(a)
cos(a)
fabs(a)=absolute value
step(a) = 0 for a<0; 1 else; (this can also be used to
calc. the maximum and minimum: Max(a,b)=a*step(a-b)+b*step(b-a) ;
Min(a,b)=a*step(b-a)+b*step(a-b) )
Comments can be added after the function definition after a '.
Examples:
((a[0]+a[1]*exp(-t/[a[2]))^2)*1e-12 ' square of a single exponential
step(1-t)*a[0] + step(t-1)*a[1] ' = a[0] for t<1 and a[1] for
t>=1
exp(a[0]+log(a[1]*sqrt(fabs(t-a[8]))))^(1-step(a[3]-t)) ' absurd
example